Jack Viljoen, a masters student in the Botany department is working on the evolution of Cape Sedges.
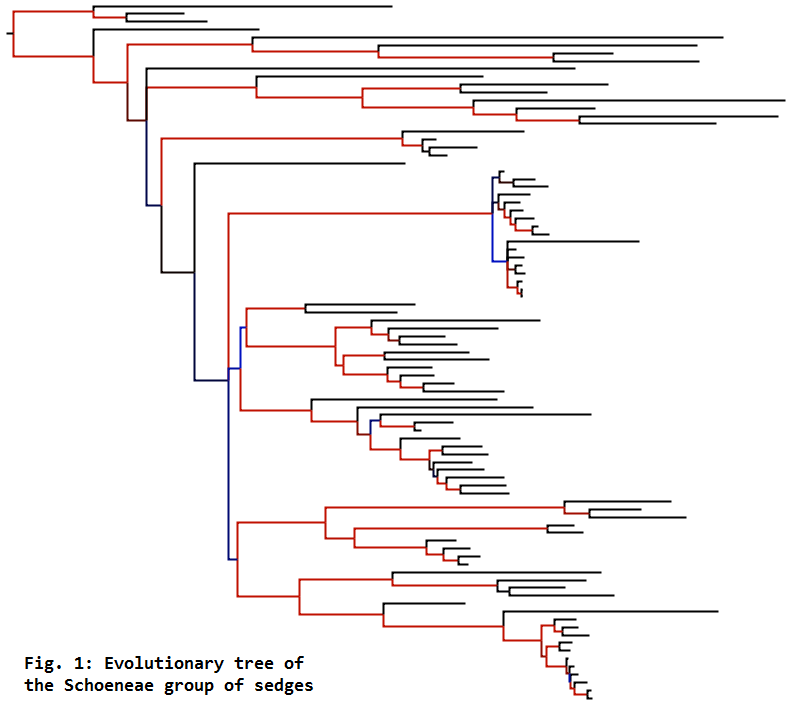
For reconstructing the evolutionary trees of groups like the Cypereae and Schoeneae (Fig. 1),
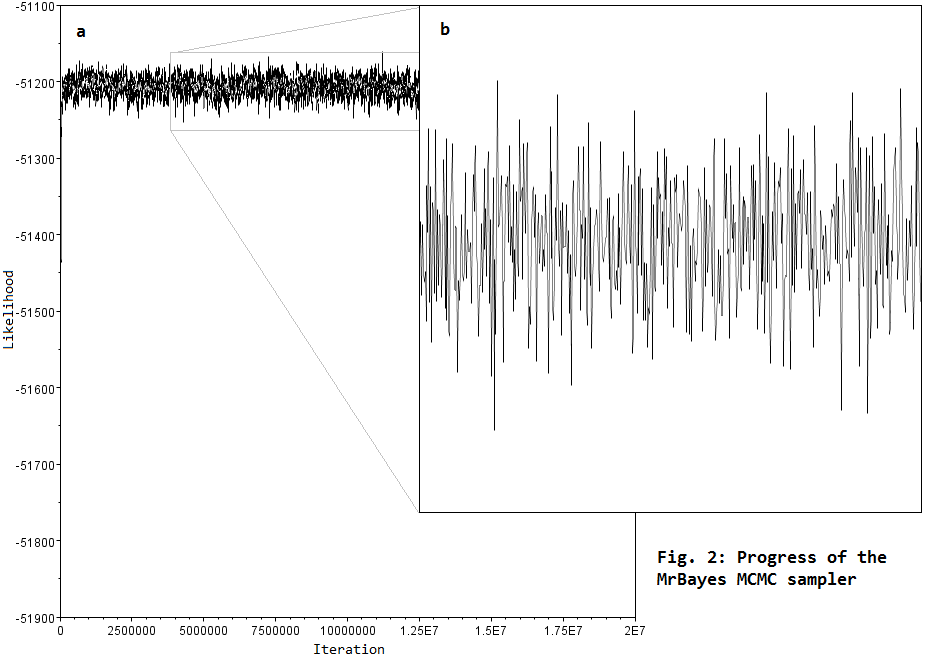
 .... we use Monte Carlo Markov Chain (MCMC) sampling to estimate parameter values related to models of DNA sequence evolution. This involves running programs like MrBayes for about 5–50 million iterations, in order to explore parameter space to find the region of highest likelihood (Fig. 2a), and to sample this region sufficiently to get good parameter estimates (Fig. 2b).
.... we use Monte Carlo Markov Chain (MCMC) sampling to estimate parameter values related to models of DNA sequence evolution. This involves running programs like MrBayes for about 5–50 million iterations, in order to explore parameter space to find the region of highest likelihood (Fig. 2a), and to sample this region sufficiently to get good parameter estimates (Fig. 2b).
 This process is made much more efficient by running multiple instances of the sampler simultaneously on different CPUs, and we are grateful to the HPC unit at UCT for allowing us to do just that.
This process is made much more efficient by running multiple instances of the sampler simultaneously on different CPUs, and we are grateful to the HPC unit at UCT for allowing us to do just that.
 .... we use Monte Carlo Markov Chain (MCMC) sampling to estimate parameter values related to models of DNA sequence evolution. This involves running programs like MrBayes for about 5–50 million iterations, in order to explore parameter space to find the region of highest likelihood (Fig. 2a), and to sample this region sufficiently to get good parameter estimates (Fig. 2b).
.... we use Monte Carlo Markov Chain (MCMC) sampling to estimate parameter values related to models of DNA sequence evolution. This involves running programs like MrBayes for about 5–50 million iterations, in order to explore parameter space to find the region of highest likelihood (Fig. 2a), and to sample this region sufficiently to get good parameter estimates (Fig. 2b).
 This process is made much more efficient by running multiple instances of the sampler simultaneously on different CPUs, and we are grateful to the HPC unit at UCT for allowing us to do just that.
This process is made much more efficient by running multiple instances of the sampler simultaneously on different CPUs, and we are grateful to the HPC unit at UCT for allowing us to do just that.