In order to test the parallel version of MrBayes we ran several
simulations of the primates.nex tutorial provided with the
installation. This compares the evolutionary species change and
divergence between various primates.
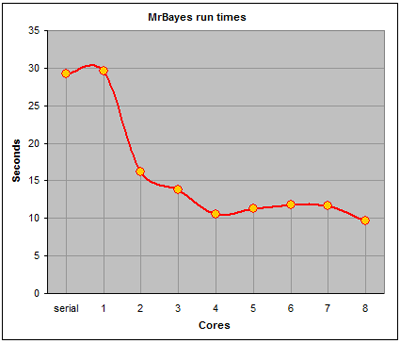
 Running the parallel version produced predictable results. "Distributing" the problem over only 1 core slowed down the computation time slightly due to the MPI overhead compared with the serial computation on only one processor. However once more cores were brought in the computation time decreased markedly until the amount of work that could be distributed was reached. This occurred at 5 cores and was in line with the documented observation that the number of chains should not exceed the number of cores. Providing more cores did not assist in computation and merely lead to more overhead in setting up MPI.
Running the parallel version produced predictable results. "Distributing" the problem over only 1 core slowed down the computation time slightly due to the MPI overhead compared with the serial computation on only one processor. However once more cores were brought in the computation time decreased markedly until the amount of work that could be distributed was reached. This occurred at 5 cores and was in line with the documented observation that the number of chains should not exceed the number of cores. Providing more cores did not assist in computation and merely lead to more overhead in setting up MPI.
 The efficiency of the cluster is related to the problem being analysed as can be seen from the documentation: "In our experience, heating is essential for problems with more than about 50 taxa, whereas smaller problems often can be analysed successfully without heating. The time complexity of the analysis is directly proportional to the number of chains used (unless MrBayes runs out of physical RAM memory, in which case the analysis will suddenly become much slower), but the cold and heated chains can be distributed among
processors in a cluster of computers using the MPI version of the
program greatly speeding up the calculations."
Hence the more data (groups of organisms) one analyses the more cores one can use in order to speed up the computation.
The efficiency of the cluster is related to the problem being analysed as can be seen from the documentation: "In our experience, heating is essential for problems with more than about 50 taxa, whereas smaller problems often can be analysed successfully without heating. The time complexity of the analysis is directly proportional to the number of chains used (unless MrBayes runs out of physical RAM memory, in which case the analysis will suddenly become much slower), but the cold and heated chains can be distributed among
processors in a cluster of computers using the MPI version of the
program greatly speeding up the calculations."
Hence the more data (groups of organisms) one analyses the more cores one can use in order to speed up the computation.
 Running the parallel version produced predictable results. "Distributing" the problem over only 1 core slowed down the computation time slightly due to the MPI overhead compared with the serial computation on only one processor. However once more cores were brought in the computation time decreased markedly until the amount of work that could be distributed was reached. This occurred at 5 cores and was in line with the documented observation that the number of chains should not exceed the number of cores. Providing more cores did not assist in computation and merely lead to more overhead in setting up MPI.
Running the parallel version produced predictable results. "Distributing" the problem over only 1 core slowed down the computation time slightly due to the MPI overhead compared with the serial computation on only one processor. However once more cores were brought in the computation time decreased markedly until the amount of work that could be distributed was reached. This occurred at 5 cores and was in line with the documented observation that the number of chains should not exceed the number of cores. Providing more cores did not assist in computation and merely lead to more overhead in setting up MPI.
 The efficiency of the cluster is related to the problem being analysed as can be seen from the documentation: "In our experience, heating is essential for problems with more than about 50 taxa, whereas smaller problems often can be analysed successfully without heating. The time complexity of the analysis is directly proportional to the number of chains used (unless MrBayes runs out of physical RAM memory, in which case the analysis will suddenly become much slower), but the cold and heated chains can be distributed among
processors in a cluster of computers using the MPI version of the
program greatly speeding up the calculations."
Hence the more data (groups of organisms) one analyses the more cores one can use in order to speed up the computation.
The efficiency of the cluster is related to the problem being analysed as can be seen from the documentation: "In our experience, heating is essential for problems with more than about 50 taxa, whereas smaller problems often can be analysed successfully without heating. The time complexity of the analysis is directly proportional to the number of chains used (unless MrBayes runs out of physical RAM memory, in which case the analysis will suddenly become much slower), but the cold and heated chains can be distributed among
processors in a cluster of computers using the MPI version of the
program greatly speeding up the calculations."
Hence the more data (groups of organisms) one analyses the more cores one can use in order to speed up the computation.